Focus on Research
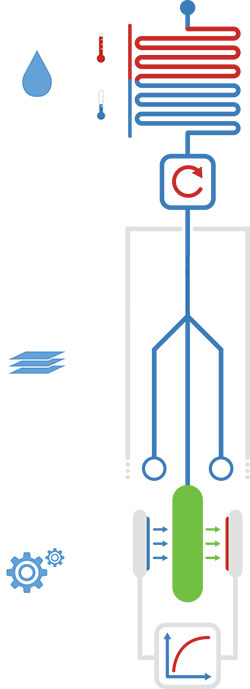
Figure 1: Overview of the three major hardware modules. Shown are, starting from above: the processing unit, the paper strip and the fluorescence detector.
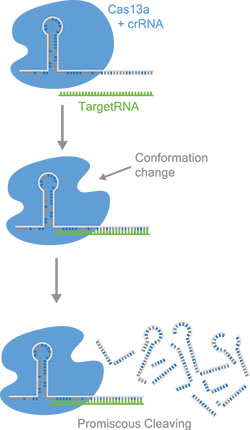
Figure 2: Cas13a binds specific target RNA depending on the crRNA sequence. After activation, Cas13a cleaves RNA indiscriminately, which is used for the detection mechanisms. The scheme vizualizes the possibilty to design new crRNAs to sense any kind of target sequence.
|
iGEM Team 2017
The Munich iGEM project CascAID+ - Cas13a controlled assay for infectious diseases
The 2017 Munich iGEM team CascAID+ has developed a highly specific diagnostic platform that differentiates between bacterial and viral infections based on the unique properties of the CRISPR protein Cas13a.
Scientific Background: The discovery of clustered regularly interspaced RNA (CRISPR) molecules in bacteria and the associated protein effectors (Cas proteins) has led to the development of a wide variety of powerful biochemical tools for the manipulation of DNA and RNA molecules. Among them, the recently discovered Cas13a has a unique mode of action. Upon binding to a CRISPR-associated RNA (crRNA), it will form a complex that can recognize an RNA molecule (the target RNA) and cut it. At the same time, Cas13a develops a collateral nuclease activity which leads to unspecific cutting of RNA molecules. The detection of the target is based on complementarity between the crRNA and the target RNA, and is highly specific (2 point mutations in the target sequence are sufficient to prevent detection).
The project: CascAID+ utilizes Cas13a by loading it with guide RNA molecules with sequences specific for bacteria or viruses. When detecting the presence of the corresponding bacterial or viral RNA, the collateral nuclease activity of Cas13a is used to generate a fluorescent signal, e.g., by cutting doubly labeled RNA. The CascAID+ team developed a complete diagnostic system around this basic detection mechanism, with an emphasis on low-cost and point of-care applications. The platform comprises a sample pre-processing unit with a portable fluidic system featuring a temperature control unit for lysis and isothermal amplification of target RNA molecules. Cas13a detection is performed on a paper strip reaction unit containing the freeze-dried detection reaction mix, which becomes active upon rehydration with the sample. The fluorescence signal generated by the reaction is read out with a low-cost portable detector unit. The team also developed software for a crRNA databank, secondary structure verification of crRNAs and off-target verification of designed crRNAs as well as for data analysis.
During the project, CascAID+ was successfully used to amplify a target from lysed cells and detect it with Cas13a. With this amplification, the platform could detect tens of attomolar concentrations of target, in the range of real sample levels. Furthermore, CascAID+ was used to detect and differentiate RNA from E. coli, Norovirus and Hepatitis C virus, three major causes of infections worldwide.
The team was led by the supervisors Aurore Dupin and Lukas Aufinger. Scientific experiments were conducted in the laboratory of Prof. Dr. Friedrich Simmel at TUM in Garching.
|
|
|
|
GRK2062 Publications
Physical Review E 96, 022408, 2017
Nonequilibrium dynamics of probe filaments in actin-myosin networks
Jannes Gladrow and Chase P. Broedersz and Christoph F. Schmidt
Abstract
Active dynamic processes of cells are largely driven by the cytoskeleton, a complex and adaptable semiflexible polymer network, motorized by mechanoenzymes. Small dimensions, confined geometries, and hierarchical structures make it challenging to probe dynamics and mechanical response of such networks. Embedded semiflexible probe polymers can serve as nonperturbing multiscale probes to detect force distributions in active polymer networks. We show here that motor-induced forces transmitted to the probe polymers are reflected in nonequilibrium bending dynamics, which we analyze in terms of spatial eigenmodes of an elastic beam under steady-state conditions. We demonstrate how these active forces induce correlations among the mode amplitudes, which furthermore break time-reversal symmetry. This leads to a breaking of detailed balance in this mode space. We derive analytical predictions for the magnitude of resulting probability currents in mode space in the white-noise limit of motor activity. We relate the structure of these currents to the spatial profile of motor-induced forces along the probe polymers and provide a general relation for observable currents on two-dimensional hyperplanes.
Full text dx. doi.org/10.1103/PhysRevE.96.022408
New Biotechnology Vol. 39, Part B, 199-205 2017
Cell-free protein synthesis in micro compartments: building a minimal cell from biobricks
Haiyang Jia, Michael Heymann, Frank Bernhard, Petra Schwille, Lei Kai
Abstract
The construction of a minimal cell that exhibits the essential characteristics of life is a great challenge in the field of synthetic biology. Assembling a minimal cell requires multidisciplinary expertise from physics, chemistry and biology. Scientists from different backgrounds tend to define the essence of ‘life’ differently and have thus proposed different artificial cell models possessing one or several essential features of living cells. Using the tools and methods of molecular biology, the bottom-up engineering of a minimal cell appears in reach. However, several challenges still remain. In particular, the integration of individual sub-systems that is required to achieve a self-reproducing cell model presents a complex optimization challenge. For example, multiple self-organisation and self-assembly processes have to be carefully tuned. We review advances and developments of new methods and techniques, for cell-free protein synthesis as well as micro-fabrication, for their potential to resolve challenges and to accelerate the development of minimal cells.
Full text http: //doi.org/10.1016/j.nbt.2017.06.014
|
|
|
|
New Members
PhD-students
 |
Christian Gebhardt, M.Sc., started his PhD study in September 2017. Supervised by Thorben Cordes he is focusing on “Mechnisms of membrane transport”. |
|
|
|
|
|
Events
|
The third Conference on Synthetic Biology will take place from July 23 - 25, 2018 at the Sparkassenakademie Landshut (about 40 km far from Munich Airport, 70 km to Munich City Center). It is one of the largest conferences in this field in Europe. The conference is planned as a platform to exchange and discuss cutting-edge results on Synthetic Biology. This symposium brings together biologists, chemists and physicists working on different aspects in Synthetic Biology.
The topics will cover all aspects ranging from mirror biology, orthogonal biosystems, metabolic and genome engineering technologies, synthetic circuits, modeling and computer-aided design as well as translational aspects of Synthetic Biology.
For GRK2062 PhD-students this conference will be the Retreat 2018.
|
|
Report by Jorge Quintero: iGEM Team 2017
When the members of these year's iGEM team were announced, around two weeks after the fist meeting, I was (happily) surprised to find my own name in the list: I had wondered whether I, with no background in synthetic biology, would make the cut. At this point, even though I still hadn't realized that iGEM is a learning opportunity as much as it is a competition, my six months as an iGEM student began.
For our project, we decided to develop a new diagnosis system using a newly characterized CRISPR/Cas effector. After intense discussion on what the name of the project should be, we chose CascAID+ and then the real work began. Primer design, protein purification, finding sponsors and Wiki design were some of our tasks during the semester. It certainly wasn't easy to balance iGEM with university courses, but I was determined on giving my best for both things. During this time I also got to know the team better: all kinds of people from different study programmes, some with a little less experience than me, some with a lot more experience, some with experience in different things. From them, I learnt new things, learnt how to approach issues through different perspectives and gained interested in topics I had never considered before. They, just like me, helped the team in doing what they knew best and picking on new things along the way. And although, like in any other team, there were some confrontations, learning how to deal with them was part of the experience. Once the semester and the exams were over, the REAL work began. I could finally focus exclusively on iGEM and spent most of my time working in the lab. I got to live the highs and lows of science, with the emotion of expected results, the thrill of unexpected results and the frustration of no results at all. I had never worked so hard or dedicated so much time in anything, but at the end, we were able to go to the Giant Jamboree proud and happy with what we had accomplished. Being there was amazing by itself, seeing what other teams had done and hear them present their projects.
To me, iGEM was a lot of work, it was fun work, frustrating work and rewarding work. But mostly, it was an amazing experience where I learnt many new skills and topics, and together with the team got the opportunity to shape and develop a project by ourselves. |
|
Group Photo:
The successful Munich iGEM Team 2017
|
|
The combined iGEM team from the two Munich universities LMU and TUM has received the 1st Runner Up Prize Overgrad in the international iGEM competition at Boston (MA) on November 13, 2017. Furthermore, CascAID+ received special awards in the category “Best Diagnostics Project”, “Best Applied Design”, “Best Hardware”, “Best Software” as well as “Best Model”. For details of the project, please refer to “Focus on Research”.
Upcoming Advanced Method Courses
PyMol Workshop
Topics: General introduction in protein structures and their analyzes as well as an introduction to the software PyMOL, which is a free and flexible molecular graphics and modelling package which can be also used to generate animated sequences.
Sabine Schneider will held this course on February 6th 2018, 3.30-6.30 pm in room CH 53 306 at the department of chemistry in Garching.
Registration: Please write an e-mail to Sabine Schneider .
Methods and concepts in protein design: Molecular Engineering in Bacteria (Jung/Lassak)
Synthetic Biology demands for efficient strategies to genetically manipulate microorganisms. In this research course we will use various molecular tools to learn how to efficiently introduce genetic material into diverse bacteria. Thus, we will focus on the three major principles of gene transfer in prokaryotes – transformation, transduction and conjugation.
The week-long course will take place from April 9-13, 2018. Registration: Please write an e-mail to GRK2062 .
|
Upcoming Transferable Skills Courses
Scientific Writing, by Brian Cusack
Participants learn how to:
- Apply five key principles of scientific writing
- Write for their readers
- Construct a memorable “take-home message”
- Connect all parts of their paper in a flowing narrative
- Overcome writers' block
- Structure their paper for increased impact
- Use the writing process to inform their own research
- Understand the role of “story telling” in scientific writing
The course will run on the 22nd and 23rd of February, 2018. Registration: Please write an e-mail to GRK2062 .
Effective Visual Communication for Scientists, by Seyens
Attention: course postponed to 2018
If created properly, graphics are the most effective way to explain complex ideas in the shortest amount of time, attract audience and raise credibility. Nevertheless, researchers aren't trained in visual communication in the traditional PhD curricula and are supposed to acquire these skills by themselves. This workshop uses a hands-on approach to help researchers visually present their own research through various means of scientific communication.
|
|
|
|
Journal Club
Nature Chemical Biology, Vol. 13, 706–708 (2017); 1651: 263-273
Brief Communication
Engineering RGB color vision into Escherichia coli
Jesus Fernandez-Rodriguez, Felix Moser, Miryoung Song and Christopher A Voigt
Optogenetic tools use colored light to rapidly control gene expression in space and time. We designed a genetically encoded system that gives Escherichia coli the ability to distinguish between red, green, and blue (RGB) light and respond by changing gene expression. We use this system to produce ‘color photographs’ on bacterial culture plates by controlling pigment production and to redirect metabolic flux by expressing CRISPRi guide RNAs.
Full text: http: //dx.doi.org/10.1038/nchembio.2390 |
|
|
|
Publisher
GRK 2062 Molecular Principles of Synthetic Biology
Ludwig-Maximilians-Universität München
LMU Biocenter
Großhaderner Str. 2-4
82152 Martinsried
Germany
Editor
Dr. Beate Hafner
Contact
E-Mail: grk2062 @bio.lmu.de
Phone: + 49 89 2180-74714
Web: http: //www.grk2062.lmu.de |
|
|