Focus on Research
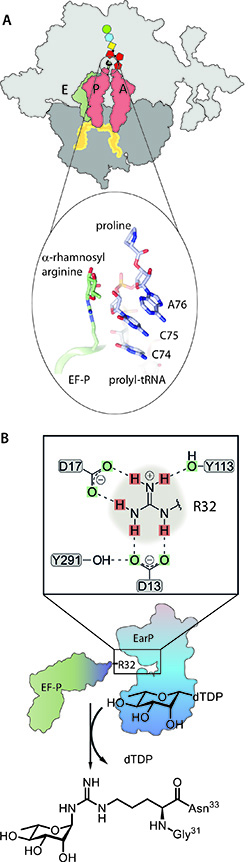
Figure 1: EF-P - Glycosylation and mode of action A EF-P binds to the diproline-locked ribosome. The interaction of α-rhamnosyl arginine with the CCA end of the P-site tRNA facilitates the formation of a new peptide bond.
B Posttranslational activation of EF-P by the glycosyltransferase EarP of P. putida with dTDP-β-L-rhamnose (TDP-Rha). Depicted is the catalytic pocket with its crucial aspartates D13 and D17, priming arginine R32 for the nucleophilic attack onto the anomeric carbon of TDP-Rha to mediate the formation of α-rhamnosyl arginine.
|
PD Dr. Jürgen Lassak
Area B Synthetic Proteins
What a bacterial translation elongation factor teaches us about glycosylation
Glycosylations are one of the most important and probably the most frequent posttranslational protein modifications in biological systems. Nevertheless, for a long time it was believed that they were limited to eukaryotes. However, bacteria also contain a large number of glycoproteins that fulfil important functions, for example in protein biosynthesis. This process comes to an unwanted stop when two or more consecutive prolines have to be translated. The evolutionary solution to this problem is a specialized protein that is recruited to the arrested ribosome and restarts the elongation of the amino acid chain (Fig. 1A). Especially important for the activity of this universal elongation factor, in bacteria EF-P, are posttranslational modifications. They play a decisive role in the stabilization and orientation of peptidyl-tRNA in order to form a new peptide bond. In pseudomonads and thus also in the pathogen of bacterial pneumonia P. aeruginosa, a conserved arginine is α-rhamnosylated by the novel glycosyltransferase EarP. Hereby the nucleotide precursor dTDP-β-L-rhamnose serves as donor substrate.
However, our discovery is not only interesting with regard to the functionalization of EF-P. On the one hand, rhamnosylation plays a crucial role for bacterial pathogenicity and thus makes EarP an interesting target structure for the development of new antimicrobial agents. On the other hand, it creates a precedent in (bacterial) glycobiology. The literature on nitrogen-linked protein glycosylations is almost exclusively limited to asparagine as acceptor amino acid and EF P is still the only known arginine glycosylated protein in bacteria. Together with a research team led by Janosch Hennig, we were recently able to solve the crystal structure of EarP and thus understand for the first time how glycosyl transfer to arginine is possible. Surprisingly, the postulated catalysis mechanism is similar to that of known asparagine glycosyltransferases (Fig. 1B). It therefore stays an open question to what extent arginine in fact remains the acceptor exception from the rule of N-linked glycosylations. Equally exciting is the examination of EF-P rhamnosylation from the sugar perspective. Just like arginine as acceptor amino acid, activated rhamnose as donor substrate appears to be rather unusual, despite its ubiquitous occurrence.
Together with Prof. Dr. Anja Hoffmann-Röder and her colleagues Daniel Gast and Swetlana Wunder, we are currently investigating the natural distribution of rhamnosylations and actually have first indications of new glycoproteins, for example in mycobacteria. In contrast to most other prokaryotes, here the dTDP-β-L-rhamnose biosynthesis pathway is essential. So far, it has been assumed that this is due to a special structure in the cell wall, the mycolic acids, which are linked to the peptidoglycan by rhamnose. Alternatively, mycobacteria may also contain an essential protein whose activity is directly linked to posttranslational glycosylation, similar to EF-P in pseudomonads. Should this be confirmed, this would perhaps be a new target for the fight against tuberculosis. Our finding also underlines once again the importance of research on bacterial glycosylations.
|
|
|
|
GRK2062 Publications
Journal of the Royal Society Interface 15, 20180495
Bacterial chromosome organization by collective dynamics of SMC condensins
Christiaan A. Miermans and Chase P. Broedersz
Abstract
A prominent organizational feature of bacterial chromosomes was revealed by Hi-C experiments, indicating anomalously high contacts between the left and right chromosomal arms. These long-range contacts have been attributed to various nucleoid-associated proteins, including the ATPase Structural Maintenance of Chromosomes (SMC) condensin. Although the molecular structure of these ATPases has been mapped in detail, it still remains unclear by which physical mechanisms they collectively generate long-range chromosomal contacts. Here, we develop a computational model that captures the subtle interplay between molecular-scale activity of slip-links and large-scale chromosome organization. We first consider a scenario in which the ATPase activity of slip-links regulates their DNA-recruitment near the origin of replication, while the slip-link dynamics is assumed to be diffusive. We find that such diffusive slip-links can collectively organize the entire chromosome into a state with aligned arms, but not within physiological constraints. However, slip-links that include motor activity are far more effective at organizing the entire chromosome over all length-scales. The persistence of motor slip-links at physiological densities can generate large, nested loops and drive them into the bulk of the DNA. Finally, our model with motor slip-links can quantitatively account for the rapid arm–arm alignment of chromosomal arms observed in vivo.
Full text https://doi.org/10.1098/rsif.2018.0495
|
|
|
|
Events
Renewal proposal GRK2062: On-site review
On January 31st and February 1st, 2019 all GRK2062 members are asked to bundle their forces to convince with an impressive overview about our research projects and teaching elements in order to get funding for the second funding period. Reviewers will especially focus on research projects of PhD candidates as well as experiences within the Research Training Group.
We count on you!
Again successful: 1st Runner Up Prize Overgrad for Munich iGEM team 2018
The successful story of the iGEM Team Munich continued. With the help of concepts from synthetic biology the team created Phactory, a cell-free molecular assembly line for bacteriophages to overcome the lack of common manufacturing procedures which meet international quality and safety standards. Their development may push forward the worldwide implementation of bacteriophage therapy - a 100-year old treatment employing the natural enemies of bacteria. Compared to traditional manufacturing procedures, Phactory requires 2.5% of the production volume and demands no special biosafety regulations to yield bacteriophages ready for therapy.
|
|
Great success in Boston:
Enthusiastic Munich iGEM Team 2018 during awards ceremony
|
|
Beside the 1st Runner Up Prize in the overgrad category, Phactory received special awards in the category “Best Manufacturing Project”, “Best Entrepreneurship”, “Best Presentation”, “Best Software” as well as “Best Wiki”.
This work wouldn’t be possible without the help of Prof. Dr. Friedrich Simmel, who hosted the iGEM team in his laboratory at the TUM in Garching, Prof. Dr. Gil Westmeyer and Prof. Dr. Kirsten Jung, who gave scientific advice, and the UnternehmerTUM, who helped with their network. Beside them the team also received financial support by the LMU, the TUM, the GRK2062 and also from industrial sponsors.
Congratulations to all members, to the supervisors as well as to the sponsors of the team!
Upcoming Transferable Skills Courses
Presentation skills by Sabine Walter and Tim Wagner
Unfortunately, only 3% of presentations captivate their audiences, while more than 80% are dull and boring.
Our aim is to ensure that your presentations are among the 3% that captivate and engage their audiences. During the 2-day workshop you will learn how to efficiently prepare a presentation and tailor it to specific audiences. You will overcome your stage fright and learn how to enjoy giving presentations. In short: you captivate your audience and win them over not only with what you are saying but also with how you are saying it.
For a full description please click here. Participation will be awarded with 1.0 ECTS.
Date: 3th and 4th of April, 2019
Registration: Please write an e-mail to GRK2062 |
|
|
|
Journal Club
Science News: Stockholm, October 3, 2018
‘Revolution based on evolution’ honored with chemistry Nobel
Three scientists who put evolution to work in the lab have won the Nobel Prize in Chemistry.
Frances H. Arnold of the California Institute of Technology in Pasadena was awarded half of the $1 million prize for her work on the “directed evolution” of enzymes, proteins that catalyze specific chemical reactions. The enzymes that resulted from her research have made it possible to develop new ways to make key pharmaceuticals and more environmentally friendly processes for making industrial chemicals. George Smith of the University of Missouri in Columbia and Gregory Winter of the Medical Research Council’s Laboratory of Molecular Biology (LMB) in Cambridge, U.K., share the other half of the award for their research on the directed evolution of antibodies, proteins the immune system uses to recognize invaders. Their findings enabled large-scale production of specific antibodies, which have made new treatments possible for autoimmune diseases, cancer, and other conditions.
“This year's prize in chemistry rewards a revolution based on evolution,” Claes Gustafsson, chair of the Nobel Committee for Chemistry, said this morning. “Our laureates have applied the principles of [Charles] Darwin in the test tubes, and used this approach to develop new types of chemicals for the greatest benefit of humankind.”
Full text |
|
|
|
Publisher
GRK 2062 Molecular Principles of Synthetic Biology
Ludwig-Maximilians-Universität München
LMU Biocenter
Großhaderner Str. 2-4
82152 Martinsried
Germany
Editor
Dr. Beate Hafner
Contact
E-Mail: grk2062 @bio.lmu.de
Phone: + 49 89 2180-74714
Web: http: //www.grk2062.lmu.de |
|
|