Focus on Research
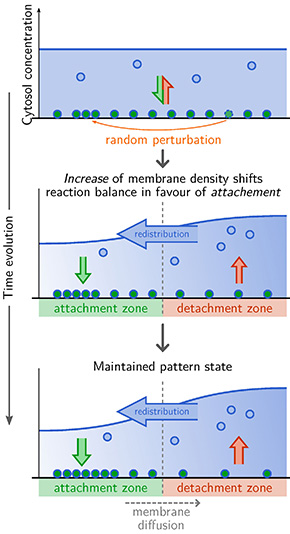
Figure 1 Linear (in)stability of a uniform initial distribution of proteins.
(a) In a uniform steady state, attachment and detachment must balance everywhere. An external cue or a random perturbation due to stochastic noise can lead a local increase in membrane density. How the relative balance of attachment and detachment processes shifts in the region of increased membrane density, determines the stability of the uniform state. If the balance in a region of increased membrane density shifts in favour of attachment (b), the region becomes an attachment zone leading to a further increase in membrane density due to redistribution through the cytosol. Hence the spatially separated attachment and detachment zones are maintained, leading the establishment of a pattern (c).
|
Prof. Dr. Erwin Frey
C2 / Area C Synthetic Systems
Self-organization principles of intracellular pattern formation
In biological systems, self-organization refers to the emergence of spatial and temporal structure. Examples include the structure of the genetic code, the structure of proteins, the structures of membrane and cytoplasm, or those of tissue, and connected neural networks. On each of these levels, interactions resulting from the dynamics and structural complementarities of the system’s constituents bring about the emergence of biological function. Biological systems are the perfect example for the Aristotelian notion that ‘the whole is more than the sum of its parts’. Owing to the advances in quantitative biology and theoretical biological physics in recent decades, we have begun to understand how biological structure and function originates from fundamental physical principles of self-organization.
Using the framework of mass-conserving reaction–diffusion equations, we systematically analyzed quantitative models for self-organized intracellular patterns, like the Min protein patterns in E. coli, Cdc42 polarization in S. cerevisiae and the bipolar PAR protein patterns found in C. elegans. It turns out that all these systems follow a common underlying principle: the formation and of spatially separated attachment and detachment zones of proteins which cycle between membrane-bound and cytosolic states. The attachment and detachment zones are coupled through cytosolic gradients that facilitate protein mass redistribution. We conclude that directed transport along an actively maintained cytosolic gradient, is the key process underlying intracellular pattern formation.
Going forward, these insights will not only help us to understand self-organization mechanisms in natural biological systems, but also provide design principles for synthetic systems able to self-organize.
Publication:
Jacob Halatek, Fridtjof Brauns and Erwin Frey: Self-organization principles of intracellular pattern formation
Philosophical Transactions of the Royal Society B 2018, April 2018
|
|
|
|
GRK2062 Publications
Science 19 Jan 2018: Vol. 359, Issue 6373
Toward dynamic structural biology: Two decades of single-molecule Förster resonance energy transfer
Eitan Lerner, Thorben Cordes, Antonino Ingargiola, Yazan Alhadid, SangYoon Chung, Xavier Michalet and Shimon Weiss
Review
Classical structural biology can only provide static snapshots of biomacromolecules. Single-molecule Förster resonance energy transfer (smFRET) paved the way for studying dynamics in macromolecular structures under biologically relevant conditions. Since its first implementation in 1996, smFRET experiments have confirmed previously hypothesized mechanisms and provided new insights into many fundamental biological processes, such as DNA maintenance and repair, transcription, translation, and membrane transport. We review 22 years of contributions of smFRET to our understanding of basic mechanisms in biochemistry, molecular biology, and structural biology. Additionally, building on current state-of-the-art implementations of smFRET, we highlight possible future directions for smFRET in applications such as biosensing, high-throughput screening, and molecular diagnostics.
Full text https://doi.org/10.1126/science.aan1133
Angew. Chem. Int. Ed. 2018, 57, 1–5
Gene Expression on DNA Biochips Patterned with Strand-Displacement Lithography
Günther Pardatscher, Matthaeus Schwarz‐Schilling, Shirley S. Daube, Roy H. Bar‐Ziv and Friedrich C. Simmel
Abstract
Lithographic patterning of DNA molecules enables spatial organization of cell‐free genetic circuits under well‐controlled experimental conditions. Here, we present a biocompatible, DNA‐based resist termed “Bephore”, which is based on commercially available components and can be patterned by both photo‐ and electron‐beam lithography. The patterning mechanism is based on cleavage of a chemically modified DNA hairpin by ultraviolet light or electrons, and a subsequent strand‐displacement reaction. All steps are performed in aqueous solution and do not require chemical development of the resist, which makes the lithographic process robust and biocompatible. Bephore is well suited for multistep lithographic processes, enabling the immobilization of different types of DNA molecules with micrometer precision. As an application, we demonstrate compartmentalized, on‐chip gene expression from three sequentially immobilized DNA templates, leading to three spatially resolved protein‐expression gradients.
Full text https://doi.org/10.1002/anie.201800281
Nano Letters March 22, 2018
Optimized Assembly of a Multifunctional RNA-Protein Nanostructure in a Cell-Free Gene Expression System
Matthaeus Schwarz-Schilling, Aurore Dupin, Fabio Chizzolini, Swati Krishnan, Sheref S. Mansy, and Friedrich C. Simmel
Abstract
Molecular complexes composed of RNA molecules and proteins are promising multifunctional nanostructures for a wide variety of applications in biological cells or in artificial cellular systems. In this study, we systematically address some of the challenges associated with the expression and assembly of such hybrid structures using cell-free gene expression systems. As a model structure, we investigated a pRNA-derived RNA scaffold functionalized with four distinct aptamers, three of which bind to proteins, streptavidin and two fluorescent proteins, while one binds the small molecule dye malachite green (MG). Using MG fluorescence and Förster resonance energy transfer (FRET) between the RNA-scaffolded proteins, we assess critical assembly parameters such as chemical stability, binding efficiency, and also resource sharing effects within the reaction compartment. We then optimize simultaneous expression and coassembly of the RNA-protein nanostructure within a single-compartment cell-free gene expression system. We demonstrate expression and assembly of the multicomponent nanostructures inside of emulsion droplets and their aptamer-mediated localization onto streptavidin-coated substrates, plus the successful assembly of the hybrid structures inside of bacterial cells.
Full text https://doi.org/10.1021/acs.nanolett.8b00526
|
|
|
|
Events
 |
|
The third Conference on Synthetic Biology will take place from July 23 - 25, 2018 in Landshut. Registration is open now. Deadline for registration is June 15th, abstract submission deadline is May 4th, 2018. GRK2062 PhD-students, PostDocs and PIs can register for free using voucher codes they have already received via e-mail. As this conference will also be the Retreat 2018, all PhD-students are obligated to present a poster or talk.
Tentative schedule
Advanced Methods Workshops
Molecular Engineering in Bacteria (Jung/Lassak)
Synthetic Biology demands for efficient strategies to genetically manipulate microorganisms. In this research course we will use various molecular tools to learn how to efficiently introduce genetic material into diverse bacteria. Thus, we will focus on the three major principles of gene transfer in prokaryotes — transformation, transduction and conjugation.
The week-long course will take place from April 9-13, 2018. Participation will be awarded with 2.5 ECTS.
The course is already fully booked.
Upcoming Transferable Skills Courses
Effective Visual Communication for Scientists, by Seyens
September 20-21, 2018
If created properly, graphics are the most effective way to explain complex ideas in the shortest amount of time, attract audience and raise credibility. Nevertheless, researchers aren't trained in visual communication in the traditional PhD curricula and are supposed to acquire these skills by themselves. This workshop uses a hands-on approach to help researchers visually present their own research through various means of scientific communication.
Participation will be awarded with 1.0 ECTS.
Registration: Please write an e-mail to GRK2062 |
|
|
|
Journal Club
Nature Vol. 538, pages 514–517 (2016)
Letter
Synchronous long-term oscillations in a synthetic gene circuit
Laurent Potvin-Trottier, Nathan D. Lord, Glenn Vinnicombe & Johan Paulsson
Synthetically engineered genetic circuits can perform a wide variety of tasks but are generally less accurate than natural systems. Here we revisit the first synthetic genetic oscillator, the repressilator1, and modify it using principles from stochastic chemistry in single cells. Specifically, we sought to reduce error propagation and information losses, not by adding control loops, but by simply removing existing features. We show that this modification created highly regular and robust oscillations. Furthermore, some streamlined circuits kept 14 generation periods over a range of growth conditions and kept phase for hundreds of generations in single cells, allowing cells in flasks and colonies to oscillate synchronously without any coupling between them. Our results suggest that even the simplest synthetic genetic networks can achieve a precision that rivals natural systems, and emphasize the importance of noise analyses for circuit design in synthetic biology.
Full text: http: //dx.doi.org/10.1038/nature19841 |
|
|
|
Publisher
GRK 2062 Molecular Principles of Synthetic Biology
Ludwig-Maximilians-Universität München
LMU Biocenter
Großhaderner Str. 2-4
82152 Martinsried
Germany
Editor
Dr. Beate Hafner
Contact
E-Mail: grk2062 @bio.lmu.de
Phone: + 49 89 2180-74714
Web: http: //www.grk2062.lmu.de |
|
|