Focus on Research
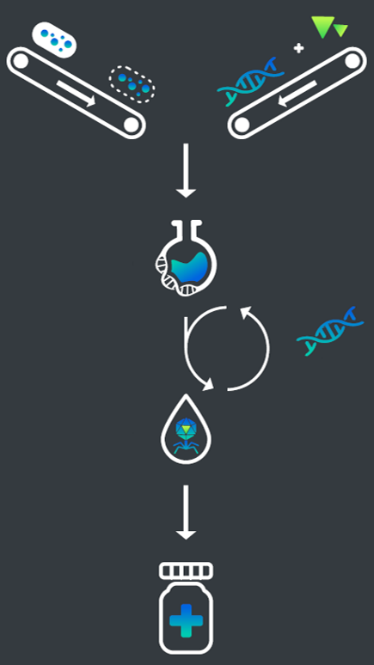
Figure 1:
Schematic overview of the cell-free bacteriophage production.
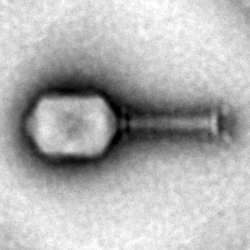
Figure 2:
TEM class averages of the EHEC phage GEC-3S assembled in cell extract.
|
Munich iGEM Team 2018
Phactory - Manufacturing Bacteriophages for Precision Medicine
The iGEM team Munich 2018, Phactory has developed a modular in vitro platform for manufacturing bacteriophages for therapeutically applications based on a cell-free E.coli extract.
Scientific Background: The emergence of antimicrobial resistances is a major threat to global health. While efforts to develop novel antimicrobial compounds have increased in recent years, there is still a lack of safe and effective substitutes for antibiotics. But there are alternatives and one of the most promising is phage therapy, which has an extensive history of successful application in countries with limited access to antibiotics. Although phage therapy has been known for almost a century, common application of phage therapy still has to become reality in the western world. What restricts phage therapy besides legal constraints are safe and clean production methods for therapeutic bacteriophages. This arises from the fact that standard bacteriophage production depends on toxic pathogenic host bacteria. Moreover, expensive and time-consuming purification methods are needed to isolate therapeutic bacteriophages from pathogenic cultures.
The project: Based on synthetic biology we created Phactory, a cell-free molecular assembly line for bacteriophages. We demonstrated the expression of several phages including T7, MS2, and GEC-3S at clinically relevant concentrations. The last is a medical relevant bacteriophage which is acting against EHEC pathogens. Exploiting the open nature of cell-free systems, Phactory enabled modular composition of bacteriophages with engineered proteins while remaining GMO-free. As a proof we assembled a modified T4 phage in the cell extract with a Hoc-YFP-fusionprotein. We also developed a quality control structure utilizing state-of-the-art bioinformatics as well as purification and encapsulation protocols. To expand our product variety while reducing cost, we optimized and engineered home-made E.coli cell-extract. Compared to traditional manufacturing procedures, Phactory requires 2.5% of the production volume and demands no special biosafety regulations to yield bacteriophages ready for therapy.
The team was led by the supervisors Thomas Frank, Elisabeth Falgenhauer, Julia Müller, Lukas Oesinghaus, Markus Joppich, Kilian Vogele and Lukas Aufinger. Scientific experiments were mainly conducted in the laboratory of Prof. Dr. Friedrich Simmel at TUM in Garching.
|
|
|
|
GRK2062 Publications
Nature Chemistry 11, 32–39 (2019)
Signalling and differentiation in emulsion-based multi-compartmentalized in vitro gene circuits
Aurore Dupin and Friedrich C. Simmel
Abstract
Multicellularity enables the growth of complex life forms as it allows for the specialization of cell types, differentiation and large-scale spatial organization. In a similar way, modular construction of synthetic multicellular systems will lead to dynamic biomimetic materials that can respond to their environment in complex ways. To achieve this goal, artificial cellular communication and developmental programs still have to be established. Here, we create geometrically controlled spatial arrangements of emulsion-based artificial cellular compartments containing synthetic in vitro gene circuitry, separated by lipid bilayer membranes. We quantitatively determine the membrane pore-dependent response of the circuits to artificial morphogen gradients, which are established via diffusion from dedicated organizer cells. Utilizing different types of feedforward and feedback in vitro gene circuits, we then implement artificial signalling and differentiation processes, demonstrating the potential for the realization of complex spatiotemporal dynamics in artificial multicellular systems.
Full text https://doi.org/10.1038/s41557-018-0174-9
ACS Synthetic Biology 8, 148–157 (2019)
Stationary patterns in a two-protein reaction-diffusion system
Philipp Glock, Beatrice Ramm, Tamara Heermann, Simon Kretschmer, Jakob Schweizer, Jonas Mücksch, Gökberk Alagöz and Petra Schwille
Abstract
Patterns formed by reaction-diffusion mechanisms are crucial for the development or sustenance of most organisms in nature. Patterns include dynamic waves, but are more often found as static distributions, such as animal skin patterns. Yet, a simplistic biological model system to reproduce and quantitatively investigate static reaction-diffusion patterns has been missing so far. Here, we demonstrate that the Escherichia coli Min system, known for its oscillatory behavior between the cell poles, is under certain conditions capable of transitioning to quasi-stationary protein distributions on membranes closely resembling Turing patterns. We systematically titrated both proteins, MinD and MinE, and found that removing all purification tags and linkers from the N-terminus of MinE was critical for static patterns to occur. At small bulk heights, dynamic patterns dominate, such as in rod-shaped microcompartments. We see implications of this work for studying pattern formation in general, but also for creating artificial gradients as downstream cues in synthetic biology applications.
Full text https://doi.org/10.1021/acssynbio.8b00415
Nano Letters 18, 7133–7140 (2018)
Light-induced printing of protein structures on membranes in vitro
Haiyang Jia, Lei Kai, Michael Heymann, Daniela A. García-Soriano, Tobias Härtel and Petra Schwille
Abstract
Reconstituting functional modules of biological systems in vitro is an important yet challenging goal of bottom-up synthetic biology, in particular with respect to their precise spatiotemporal regulation. One of the most desirable external control parameters for the engineering of biological systems is visible light, owing to its specificity and ease of defined application in space and time. Here we engineered the PhyB-PIF6 system to spatiotemporally target proteins by light onto model membranes and thus sequentially guide protein pattern formation and structural assembly in vitro from the bottom up. We show that complex micrometer-sized protein patterns can be printed on time scales of seconds, and the pattern density can be precisely controlled by protein concentration, laser power, and activation time. Moreover, when printing self-assembling proteins such as the bacterial cytoskeleton protein FtsZ, the targeted assembly into filaments and large-scale structures such as artificial rings can be accomplished. Thus, light mediated sequential protein assembly in cell-free systems represents a promising approach to hierarchically building up the next level of complexity toward a minimal cell.
Full text https://doi.org/10.1021/acs.nanolett.8b03187
|
|
|
|
Events
Joint Retreat 2019 - ICMSE Pre-conference
Date: August 17-18, 2019
Location: Stadsschouwburg, city center of Nijmegen
We will soon send information about registration for the pre- and main-conference (ICMSE 2019). GRK2062 will organize your travel to Nijmegen on August 16, 2019 and back to Munich on August 20, 2019.
The pre-conference is jointly organised by fellows of the NCCR Molecular Systems Engeneering, as well as FMS Research Center and students of our Research Training Group GRK2062 “Molecular Principles of Synthetic Biology”.
|
|

|
iGEM 2019: Lab of Gil Westmeyer will host this year's iGEM team
Gil Westmeyer has taken on responsibility for this year's iGEM team. He follows Arne Skerra (2016) and Friedrich Simmel (2017, Westmeyer lab at HelmholtzZentrum München
Upcoming Transferable Skills Courses
Presentation skills by Sabine Walter and Tim Wagner
Unfortunately, only 3% of presentations captivate their audiences, while more than 80% are dull and boring.
Our aim is to ensure that your presentations are among the 3% that captivate and engage their audiences. During the 2-day workshop you will learn how to efficiently prepare a presentation and tailor it to specific audiences. You will overcome your stage fright and learn how to enjoy giving presentations. In short: you captivate your audience and win them over not only with what you are saying but also with how you are saying it.
For a full description please click here. Participation will be awarded with 1.0 ECTS.
Date: 3th and 4th of April, 2019
Registration: Please write an e-mail to GRK2062
Adobe Illustrator
Andreas Binder will hold this course on the 15th and 16th of July 2019 in room G00.037 at LMU Biocenter. The workshop will cover both basic and more advanced functions of the software, which will be helpful to generate scientific posters, figures and presentations (more details). Registration: Please send an e-mail to GRK2062 |
|
|
|
Journal Club
Science 363, 884-887 (2019)
Hachimoji DNA and RNA: A genetic system with eight building blocks
Shuichi Hoshika, Nicole A. Leal, Myong-Jung Kim1, Myong-Sang Kim, Nilesh B. Karalkar, Hyo-Joong Kim, Alison M. Bates, Norman E. Watkins Jr., Holly A. SantaLucia, Adam J. Meyer, Saurja DasGupta, Joseph A. Piccirilli, Andrew D. Ellington, John SantaLucia Jr., Millie M. Georgiadis and Steven A. Benner
We report DNA- and RNA-like systems built from eight nucleotide “letters” (hence the name “hachimoji”) that form four orthogonal pairs. These synthetic systems meet the structural requirements needed to support Darwinian evolution, including a polyelectrolyte backbone, predictable thermodynamic stability, and stereoregular building blocks that fit a Schrödinger aperiodic crystal. Measured thermodynamic parameters predict the stability of hachimoji duplexes, allowing hachimoji DNA to increase the information density of natural terran DNA. Three crystal structures show that the synthetic building blocks do not perturb the aperiodic crystal seen in the DNA double helix. Hachimoji DNA was then transcribed to give hachimoji RNA in the form of a functioning fluorescent hachimoji aptamer. These results expand the scope of molecular structures that might support life, including life throughout the cosmos.
https://doi.org/10.1126/science.aat0971 |
|
|
|
Publisher
GRK 2062 Molecular Principles of Synthetic Biology
Ludwig-Maximilians-Universität München
LMU Biocenter
Großhaderner Str. 2-4
82152 Martinsried
Germany
Editor
Dr. Beate Hafner
Contact
E-Mail: grk2062 @bio.lmu.de
Phone: + 49 89 2180-74714
Web: http: //www.grk2062.lmu.de |
|
|