Focus on Research
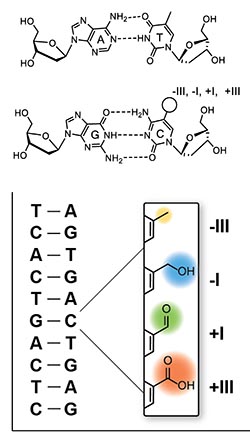
Figure 1 Depiction of the four canonical Watson-Crick base bases and their base pairs in DNA.
The bottom pictures shows how the C5-position of the DNA base dC is modified which generates new non-canonical bases that are needed to regulate transcriptional activity.
|
Prof. Dr. Thomas Carell
B1 / Area B Synthetic Proteins
Synthetic Biological Chemistry
The Carell group is investigating non-canonical nucleobases in DNA and RNA.[1] These nucleic acids are central molecules of life that are composed out of four canonical bases {(d)A, (d)C, (d)G and dT/U). The sequence of these canonical bases establish the sequence information layer that encodes life. It is this sequence information that is translated into a peptide sequence for folding into a functional protein. DNA and RNA contain next to these four canonical bases a number of partially heavily modified non-canonical bases. In RNA more than 120 modified bases have been discovered so far.[2] These bases are required for proper folding of the RNA molecule into a defined three dimensional structure and in tRNA, those present in the anticodon loop are needed to fine tune the translational process. Modified bases exist also in DNA and here particularly the cytosine bases are often modified. [1] Next to cytosine itself (Fig. 1), the genetic material contains 5-methylcytosine, 5-hydroxymethylcytosine, 5-formylcytosine and 5-carboxylcytosine in which a C-atom in the C5 position features different oxidations states ranging from -III to +I. These modified dC-bases seem to be essential for the regulation of transcription activity.
The Carell group is synthesizing modified DNA and RNA bases and to this end they develop synthetic methods. The modified bases are prepared as phosphoramidites, which allows to incorporate them into defined DNA/RNA sequences. To this end new solid phase synthesis techniques are invented. With these oligonucleotides in hand, the group investigates the function and the biological consequences of individual non-canonical bases. High end mass spectrometry is used to find the proteins that make, read and erase these bases. By knowing the identity of specific reader proteins one can deduce biological functions. Finally, metabolic feeding experiments with synthetic nucleoside analogs is performed to study how modified bases are chemically further altered in the cell. Metabolic feeding studies [3] with synthetic stable isotope labelled nucleosides as antimetabolites allows precise investigation of the chemistry that occurs in live cells on these bases. The final aim of our research is to decode the information layer beyond the sequence code in order to get a better understanding of how the genetic system operates. To this end we combine synthetic methods with cell biology and analytical chemistry.
[1] see GRK publications
[2] Thomas Carell et al. Angew. Chemie. Int. Ed. 2012, 51
Structure and Function of Noncanonical Nucleobases
[3] For an example of a metabolic labelling study see:
Katharina Iwan et al. Nat. Chem. Biol. 2017
5-Formylcytosine to cytosine conversion by C-C bond cleavage in vivo
|
|
|
|
GRK2062 Publications
Angew. Chem. Int. Ed. 10.1002/anie.201712002
Optical control of protein pattern formation
Philipp Glock, Johannes Broichhagen, Simon Kretschmer, Philipp Blumhardt, Jonas Mücksch, Dirk Trauner and Petra Schwille
Abstract
Patterns formed by protein reactions and diffusion are the foundation for many phenomena in biology. Yet, the experimental study of reaction-diffusion (R-D) systems has so far been dominated by chemical oscillators, for which many manipulation tools are available. Here, we developed a photoswitch for the Min system of Escherichia coli, a versatile biological in vitro R-D system consisting of the antagonistic proteins MinD and MinE. A MinE-derived peptide of 19 amino acids is covalently modified with a photoisomerizable crosslinker based on azobenzene to externally control peptide-mediated depletion of MinD from the membrane. In addition to providing an on-off switch for pattern formation, we achieve frequency-locked entrainment with a precise 2D spatial memory, allowing new insights into Min protein action on the membrane. Taken together, we provide a tool to externally control protein patterns formed by self-organization.
Full text dx. doi.org/10.1002/anie.201712002
Angew. Chem. Int. Ed. 10.1002/anie.201708228
Non-canonical bases in the genome: The regulatory information layer in DNA
Thomas Carell, Matthias Q Kurz, Markus Müller, Martin
Rossa, and Fabio Spada
Review
The sequence of the four canonical bases dA, dC, dG and dT forming two defined Watson-Crick base pairs (dT:dA, dG:dC), held together by H-bonding, establish the sequence information in the DNA double strand. The faithful replication of the sequence information during cell division, the transcription of the DNA information into RNA and the final translation of the sequence information into proteins is the basis for life on earth. Multicellular organisms developed the concept of specialized cells that perform specific functions. Examples are neurons and fibroblast to name just two out of more than 200. These cellular differences are established based on the same sequence information stored in the cell nucleus of all cells of an organism. The sequence information needs consequently different interpretations by the different cell types. During cellular development, when a zygote develops finally into a complex organism with its multitude of specialized cells, this interpretation of the genetic code has to be tightly regulated in space and time. Interpretation of the sequence information involves the controlled activation and silencing of specific genes so that certain proteins are made in one cell type but not in others. This involves an additional regulatory information layer beyond the pure base sequence. One aspect of this regulatory information layer relies on functional groups that are attached to the C(5) position of the canonical base dC. Currently four regulatory, non-canonical bases with a methyl (CH3)-, a hydroxymethyl (CH2OH)-, a formyl (CHO)- and a carboxyl (COOH)- group are known. While 5-methyl-cytidine is long recognized to be a regulatory base in the genome, the other three bases and the enzymes responsible for generating them, were just recently discovered. This review summarizes the discovery of the new bases and focusses at the chemical biology aspects associated with the regulatory DNA bases beyond Watson and Crick.
Full text http: //doi.org/10.1002/anie.201708228
|
|
|
|
Events
 |
|
The third Conference on Synthetic Biology will take place from July 23 - 25, 2018 in Landshut. Registration will be opened by March 15, 2018. GRK2062 PhD-students and PIs will get voucher codes for free registration.
Confirmed speakers:
- Naama Barkai, Weizman Institute, Israel
- Roy Bar-Ziv, Weizman Institute, Israel
- Nediljko Budisa, TU Berlin, Germany
- Simon Elsässer, Karolinska Institutet, Sweden
- Tobias Erb, MPI Marburg, Germany
- Nikta Fakhri, MIT, USA
- Neil St. John Forbes, University of Massachusetts, USA
- Michael Hecht, Princeton University, USA
- Poul Erik Jensen, University of Copenhagen, Denmark
- Liedewij Laan, TU Delft, The Netherlands
- Kathrin Lang, TU Munich, Germany
- Joseph Loparo, Harvard Medical School, USA
- James Murray, Imperial College London, UK
- Pamela Silver, Harvard Medical School, USA
- Jörg Stülke, Universität Göttingen, Germany
- Eriko Takano, University of Manchester, UK
- Chris Voigt, MIT, USA
- Christoph Wittmann, Universität des Saarlandes, Germany
Panel discussion: Experiences as postdoctoral fellow
Interested in staying in Academia?
Four postdoctoral fellows all with different backgrounds (biology, biophysics, biotechnology, pharmacy) will share their personal experiences with the audience.
The panel will be moderated by Kirsten Jung.
Date: March 19th, 5 pm
Location: Lecture hall G00.001 at LMU Biocenter
Speakers list:
Advanced Methods Workshops
PyMol Workshop
Topics: General introduction in protein structures and their analyzes as well as an introduction to the software PyMOL, which is a free and flexible molecular graphics and modelling package which can be also used to generate animated sequences.
Sabine Schneider will hold this course on February 6th 2018, 3.30-6.30 pm in room CH 53 306 at the department of chemistry in Garching.
Registration: Please write an e-mail to Sabine Schneider.
Methods and concepts in protein design: Molecular Engineering in Bacteria (Jung/Lassak)
Synthetic Biology demands for efficient strategies to genetically manipulate microorganisms. In this research course we will use various molecular tools to learn how to efficiently introduce genetic material into diverse bacteria. Thus, we will focus on the three major principles of gene transfer in prokaryotes — transformation, transduction and conjugation.
The week-long course will take place from April 9-13, 2018. Participation will be awarded with 2.5 ECTS.
Registration: Please write an e-mail to GRK2062 .
Upcoming Transferable Skills Courses
Scientific Writing, by Brian Cusack
February 22-23, 2018 Course contents
Room G00.031, LMU Biocenter
Participation will be awarded with 1.0 ECTS.
Effective Visual Communication for Scientists, by Seyens
September 20-21, 2018
If created properly, graphics are the most effective way to explain complex ideas in the shortest amount of time, attract audience and raise credibility. Nevertheless, researchers aren't trained in visual communication in the traditional PhD curricula and are supposed to acquire these skills by themselves. This workshop uses a hands-on approach to help researchers visually present their own research through various means of scientific communication.
Participation will be awarded with 1.0 ECTS.
Registration: Please write an e-mail to GRK2062 |
|
|
|
Journal Club
Nature Communications 9, 64 (2018)
Brief Communication
Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors
Ye Chen, Joanne M. L. Ho, David L. Shis, Chinmaya Gupta, James Long, Daniel S. Wagner, William Ott, Krešimir Josić and Matthew R. Bennett
One challenge for synthetic biologists is the predictable tuning of genetic circuit regulatory components to elicit desired outputs. Gene expression driven by ligand-inducible transcription factor systems must exhibit the correct ON and OFF characteristics: appropriate activation and leakiness in the presence and absence of inducer, respectively. However, the dynamic range of a promoter (i.e., absolute difference between ON and OFF states) is difficult to control. We report a method that tunes the dynamic range of ligand-inducible promoters to achieve desired ON and OFF characteristics. We build combinatorial sets of AraC-and LasR-regulated promoters containing -10 and -35 sites from synthetic and Escherichia coli promoters. Four sequence combinations with diverse dynamic ranges were chosen to build multi-input transcriptional logic gates regulated by two and three ligand-inducible transcription factors (LacI, TetR, AraC, XylS, RhlR, LasR, and LuxR). This work enables predictable control over the dynamic range of regulatory components.
Full text: http: //dx.doi.org/10.1038/s41467-017-02473-5 |
|
|
|
Publisher
GRK 2062 Molecular Principles of Synthetic Biology
Ludwig-Maximilians-Universität München
LMU Biocenter
Großhaderner Str. 2-4
82152 Martinsried
Germany
Editor
Dr. Beate Hafner
Contact
E-Mail: grk2062 @bio.lmu.de
Phone: + 49 89 2180-74714
Web: http: //www.grk2062.lmu.de |
|
|